Phaeodactylum

Diatoms are a ubiquitous class of microalgae being particularly important for biogeochemical cycling of minerals, such as silica, and for global carbon fixation (Falkowski et al, 19986).
The most well-known feature of diatoms is their highly ornamented external cell wall made of amorphous silica, called the frustule (Armbrust, 20091). Phaeodactylum tricornutum, although only partially silicified and considered of limited ecological relevance, has become one of the key organisms to study diatoms biology because of its ability to grow under sterile and easy conditions, the lack of a size reduction after mitotic division, high division rates allowing a fast biomass accumulation, the known morphology and the feasibility of cryogenic preservation. P. tricornutum is the second diatom for which a whole genome sequence has been generated (Bowler et al, 20082) and it is the most established species for molecular- and cellular-based studies. A series of genetic resources are available for this species (e.g., genetic transformation, EST databases, gateway vectors, gene silencing).
Phaeodactylum tricornutum
Lineage (full)
Eukaryota; Stramenopiles; Bacillariophyta; Bacillariophyceae; Bacillariophycidae; Naviculales; Phaeodactylaceae; Phaeodactylum; Phaeodactylum tricornutum
The P. tricornutum clone used for the whole genome sequencing and for the generation of our genetic resources is the CCAP1055/1, available from CCAP. It derived a from monoclonal culture of a single fusiform cell from strain CCMP632. The CCMP632 strain was originally isolated in 1956 off Blackpool (UK). P. tricornutum CCAP1055/1 has been maintained in culture continuously in F/2 medium (Guillard et al, 19757).
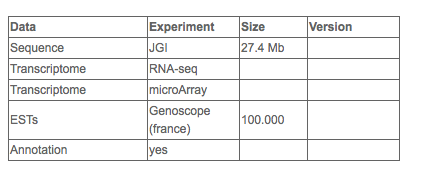
Genetic resources
Available individuals
In addition to the P. tricornutum strain used for the genome sequence and generation of genetic resources, several ecotypes are available, isolated from different geographical regions and with differential capacities to adapt to diverse conditions (De Martino et al, 20073).
Toolbox
Complete genome sequence (Bowler et al, 20082) (http://jgi.doe.gov/phaeodactylum)
Extensive Transcriptome Data (ESTs, RNA seq data, small RNA sequence data, whole genome methylome, microarray data)
Vectors for gene silencing (De Riso et al, 20094)
Gateway vectors for gene over-expression (Siaut et al, 200710)
Selected methods:
- Genetic transformation - Helium-accelerated particle bombardment (Falciatore et al, 19995)
- Gene Silencing (De Riso et al, 20094)
- Gain of Function Mutants by Activation tagging (Assemble project, SZN Naples, IT, Russo M, in progress)
- Cryogenic preservation (Falciatore et al, 19995)
- ESTs and EST database (http://www.diatomics.biologie.ens.fr/EST/) (Montsant et al, 20059; Maheswari et al, 20058)
Culture:
Phaeodactylum tricornutum is grown in F/2 medium. (Guillard et al, 19757)
Contact:
Angela Falciatore, Biologie computationnelle et quantitative, UMR7238, CNRS-UPMC. Equipe de Génomique Fonctionnelle des Diatomées, campus des Cordeliers, 15 Rue de l’Ecole de Médecine, 75006, Paris, France (www.lgm.upmc.fr/DiatomGenomics).
External links:
JGI http://genome.jgi-psf.org/Phatr2/Phatr2.home.html
EST Database http://www.diatomics.biologie.ens.fr/EST/
CCAP http://www.ccap.ac.uk/
References:
- Armbrust EV.(2009). The life of diatoms in the world's oceans. Nature, 14; 459 (7244): 185-92.
- Bowler C et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature, 13; 456 (7219): 239-44.
- De Martino A, Meichenin A, Shi J, Pan K, Bowler C (2007). Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. Journal of Phycology, 43, 992–1009
- De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A (2009) Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Research, 37 (14).
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C (1999) Transformation of Nonselectable Reporter Genes in Marine Diatoms. Marine Biotechnology,1 (3): 239-251.
- Falkowski PG, Barber RT, Smetacek VV (1998) Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science, 10; 281 (5374): 200-7.
- Guillard RRL (1975). Culture of phytoplankton for feeding marine invertebrates. Culture of Marine Invertebrate Animals, 29-60.
- Maheswari U, Montsant A, Goll J, Krishnasamy S, Rajyashri KR, Patell VM, Bowler C (2005). The Diatom EST Database. Nucleic Acids Research, 1 ;33.
- Montsant A, Jabbari K, Maheswari U, Bowler C (2005). Comparative genomics of the pennate diatom Phaeodactylum tricornutum. Plant Physiology, 137 (2): 500-13.
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene, 30; 406 (1-2): 23-35.